Bayesian graph convolutional neural networks for semi-supervised classification
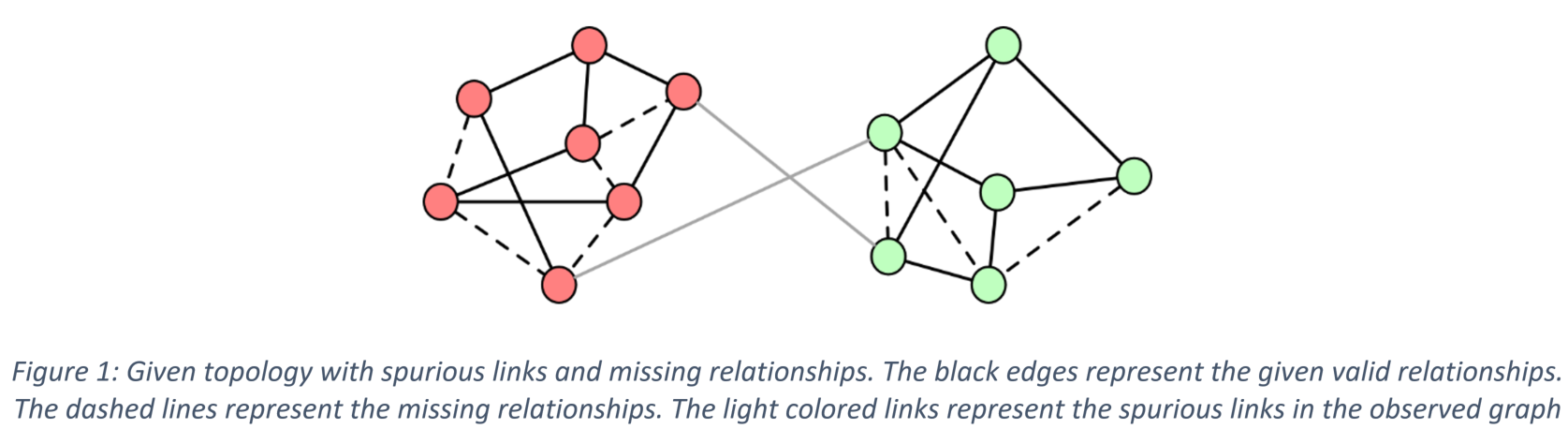
Recently, techniques for applying convolutional neural networks to graph-structured data have emerged. Graph convolutional neural networks (GCNNs) have been used to address node and graph classification and matrix completion. Although the performance has been impressive, the current implementations have limited capability to incorporate uncertainty in the graph structure. Almost all GCNNs process a graph as though it is a ground-truth depiction of the relationship between nodes, but often the graphs employed in applications are themselves derived from noisy data or modelling assumptions. Spurious edges may be included; other edges may be missing between nodes that have very strong relationships. In this paper we adopt a Bayesian approach, viewing the observed graph as a realization from a parametric family of random graphs. We then target inference of the joint posterior of the random graph parameters and the node (or graph) labels. We present the Bayesian GCNN framework and develop an iterative learning procedure for the case of assortative mixed-membership stochastic block models. We present the results of experiments that demonstrate that the Bayesian formulation can provide better performance when there are very few labels available during the training process.
PDF Abstract