Truly Generalizable Radiograph Segmentation with Conditional Domain Adaptation
Digitization techniques for biomedical images yield different visual patterns in radiological exams. These differences may hamper the use of data-driven approaches for inference over these images, such as Deep Neural Networks. Another noticeable difficulty in this field is the lack of labeled data, even though in many cases there is an abundance of unlabeled data available. Therefore an important step in improving the generalization capabilities of these methods is to perform Unsupervised and Semi-Supervised Domain Adaptation between different datasets of biomedical images. In order to tackle this problem, in this work we propose an Unsupervised and Semi-Supervised Domain Adaptation method for segmentation of biomedical images using Generative Adversarial Networks for Unsupervised Image Translation. We merge these unsupervised networks with supervised deep semantic segmentation architectures in order to create a semi-supervised method capable of learning from both unlabeled and labeled data, whenever labeling is available. We compare our method using several domains, datasets, segmentation tasks and traditional baselines, such as unsupervised distance-based methods and reusing pretrained models both with and without Fine-tuning. We perform both quantitative and qualitative analysis of the proposed method and baselines in the distinct scenarios considered in our experimental evaluation. The proposed method shows consistently better results than the baselines in scarce labeled data scenarios, achieving Jaccard values greater than 0.9 and good segmentation quality in most tasks. Unsupervised Domain Adaptation results were observed to be close to the Fully Supervised Domain Adaptation used in the traditional procedure of Fine-tuning pretrained networks.
PDF Abstract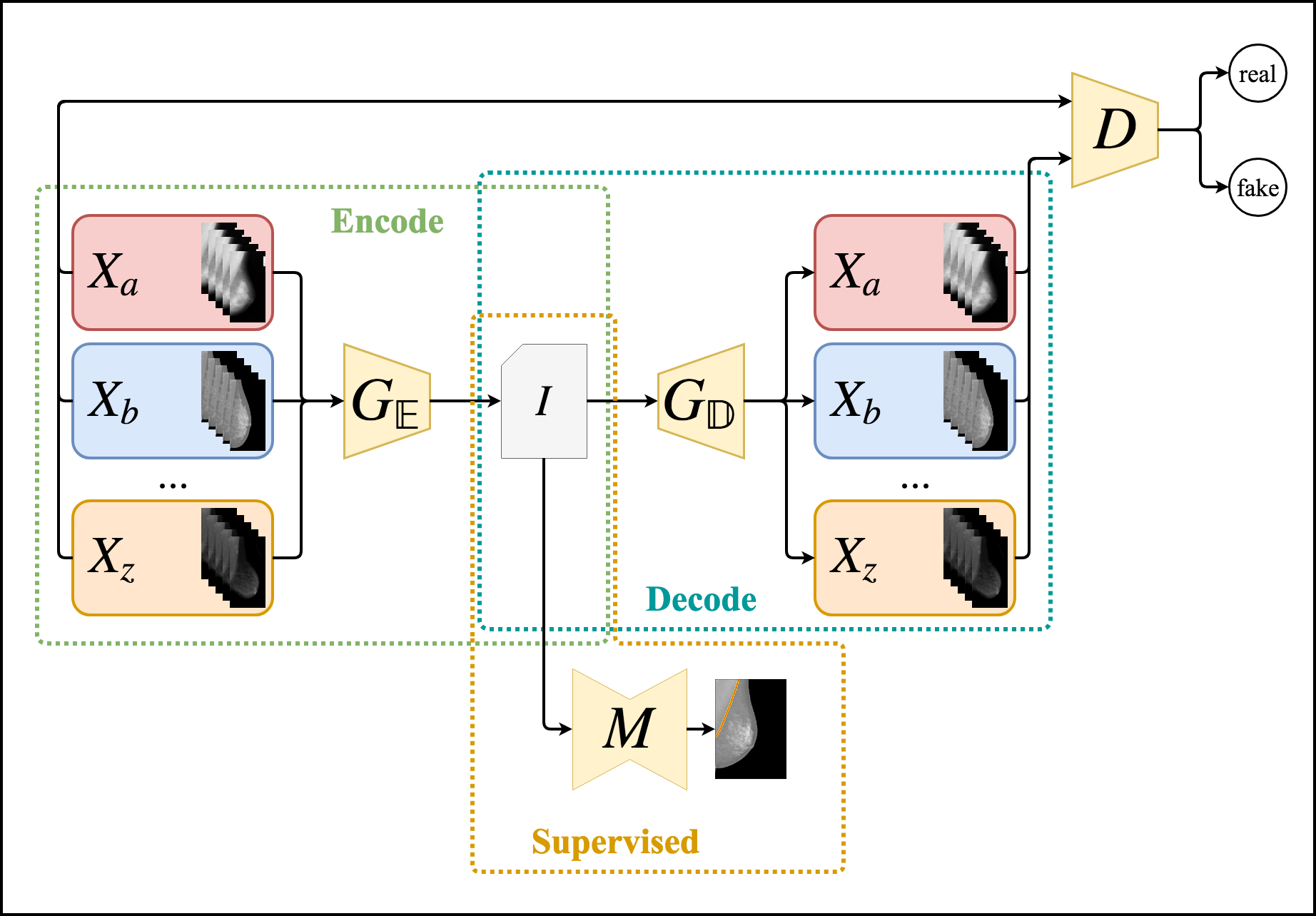






 ImageNet
ImageNet
 MNIST
MNIST
 Cityscapes
Cityscapes
 SYNTHIA
SYNTHIA
 USPS
USPS
 GTA5
GTA5
 PadChest
PadChest