Dynamic Neural Fields for Learning Atlases of 4D Fetal MRI Time-series
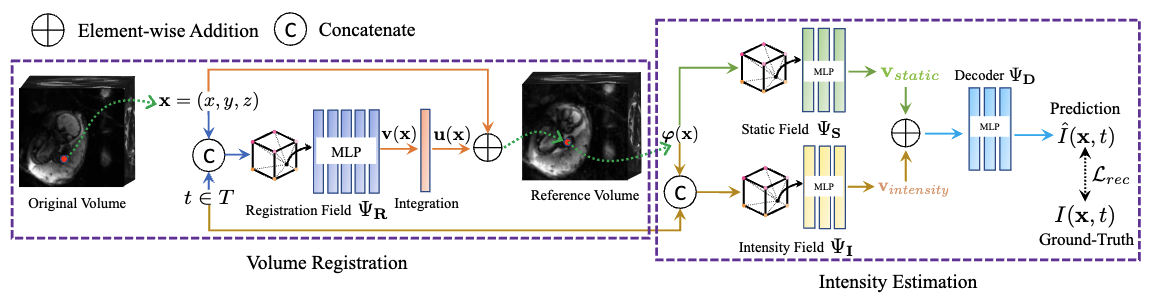
We present a method for fast biomedical image atlas construction using neural fields. Atlases are key to biomedical image analysis tasks, yet conventional and deep network estimation methods remain time-intensive. In this preliminary work, we frame subject-specific atlas building as learning a neural field of deformable spatiotemporal observations. We apply our method to learning subject-specific atlases and motion stabilization of dynamic BOLD MRI time-series of fetuses in utero. Our method yields high-quality atlases of fetal BOLD time-series with $\sim$5-7$\times$ faster convergence compared to existing work. While our method slightly underperforms well-tuned baselines in terms of anatomical overlap, it estimates templates significantly faster, thus enabling rapid processing and stabilization of large databases of 4D dynamic MRI acquisitions. Code is available at https://github.com/Kidrauh/neural-atlasing
PDF Abstract