Efficient 3D Fully Convolutional Networks for Pulmonary Lobe Segmentation in CT Images
The human lung is a complex respiratory organ, consisting of five distinct anatomic compartments called lobes. Accurate and automatic segmentation of these pulmonary lobes from computed tomography (CT) images is of clinical importance for lung disease assessment and treatment planning. However, this task is challenging due to ambiguous lobar boundaries, anatomical variations and pathological deformations. In this paper, we propose a high-resolution and efficient 3D fully convolutional network to automatically segment the lobes. We refer to the network as Pulmonary Lobe Segmentation Network (PLS-Net), which is designed to efficiently exploit 3D spatial and contextual information from high-resolution volumetric CT images for effective volume-to-volume learning and inference. The PLS-Net is based on an asymmetric encoder-decoder architecture with three novel components: (i) 3D depthwise separable convolutions to improve the network efficiency by factorising each regular 3D convolution into two simpler operations; (ii) dilated residual dense blocks to efficiently expand the receptive field of the network and aggregate multi-scale contextual information for segmentation; and (iii) input reinforcement at each downsampled resolution to compensate for the loss of spatial information due to convolutional and downsampling operations. We evaluated the proposed PLS-Net on a multi-institutional dataset that consists of 210 CT images acquired from patients with a wide range of lung abnormalities. Experimental results show that our PLS-Net achieves state-of-the-art performance with better computational efficiency. Further experiments confirm the effectiveness of each novel component of the PLS-Net.
PDF Abstract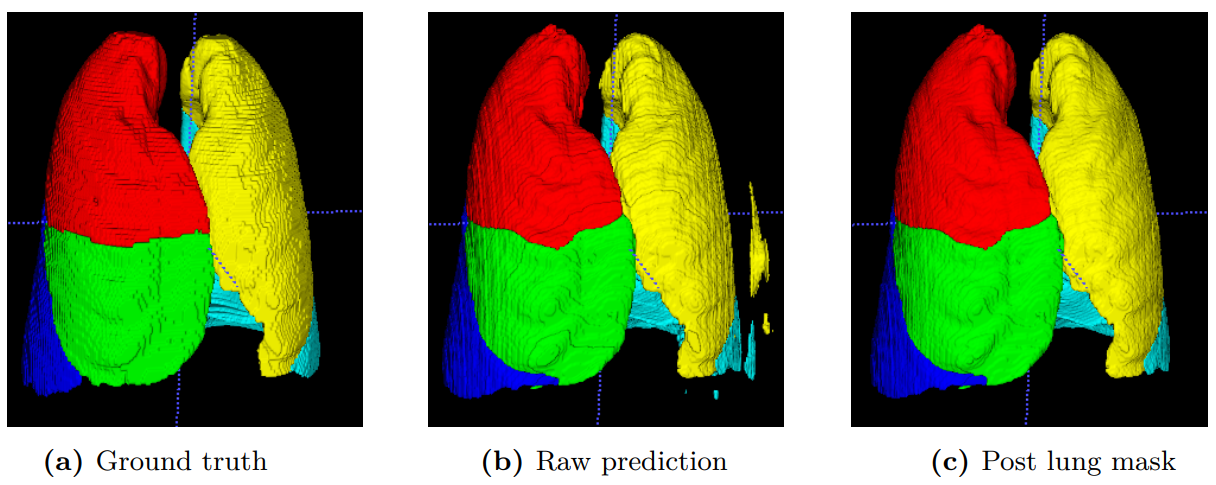



 LIDC-IDRI
LIDC-IDRI