N-Gram Graph: Simple Unsupervised Representation for Graphs, with Applications to Molecules
Machine learning techniques have recently been adopted in various applications in medicine, biology, chemistry, and material engineering. An important task is to predict the properties of molecules, which serves as the main subroutine in many downstream applications such as virtual screening and drug design. Despite the increasing interest, the key challenge is to construct proper representations of molecules for learning algorithms. This paper introduces the N-gram graph, a simple unsupervised representation for molecules. The method first embeds the vertices in the molecule graph. It then constructs a compact representation for the graph by assembling the vertex embeddings in short walks in the graph, which we show is equivalent to a simple graph neural network that needs no training. The representations can thus be efficiently computed and then used with supervised learning methods for prediction. Experiments on 60 tasks from 10 benchmark datasets demonstrate its advantages over both popular graph neural networks and traditional representation methods. This is complemented by theoretical analysis showing its strong representation and prediction power.
PDF Abstract NeurIPS 2019 PDF NeurIPS 2019 Abstract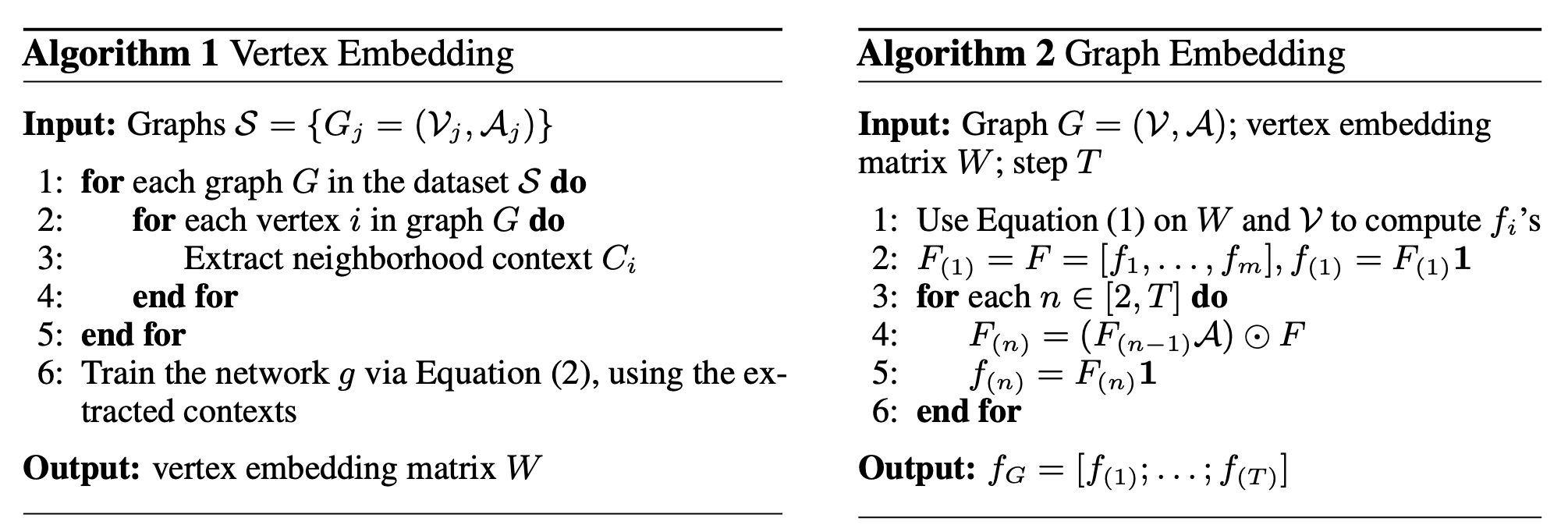












 QM9
QM9
 Tox21
Tox21
 SIDER
SIDER
