Rapid detection and recognition of whole brain activity in a freely behaving Caenorhabditis elegans
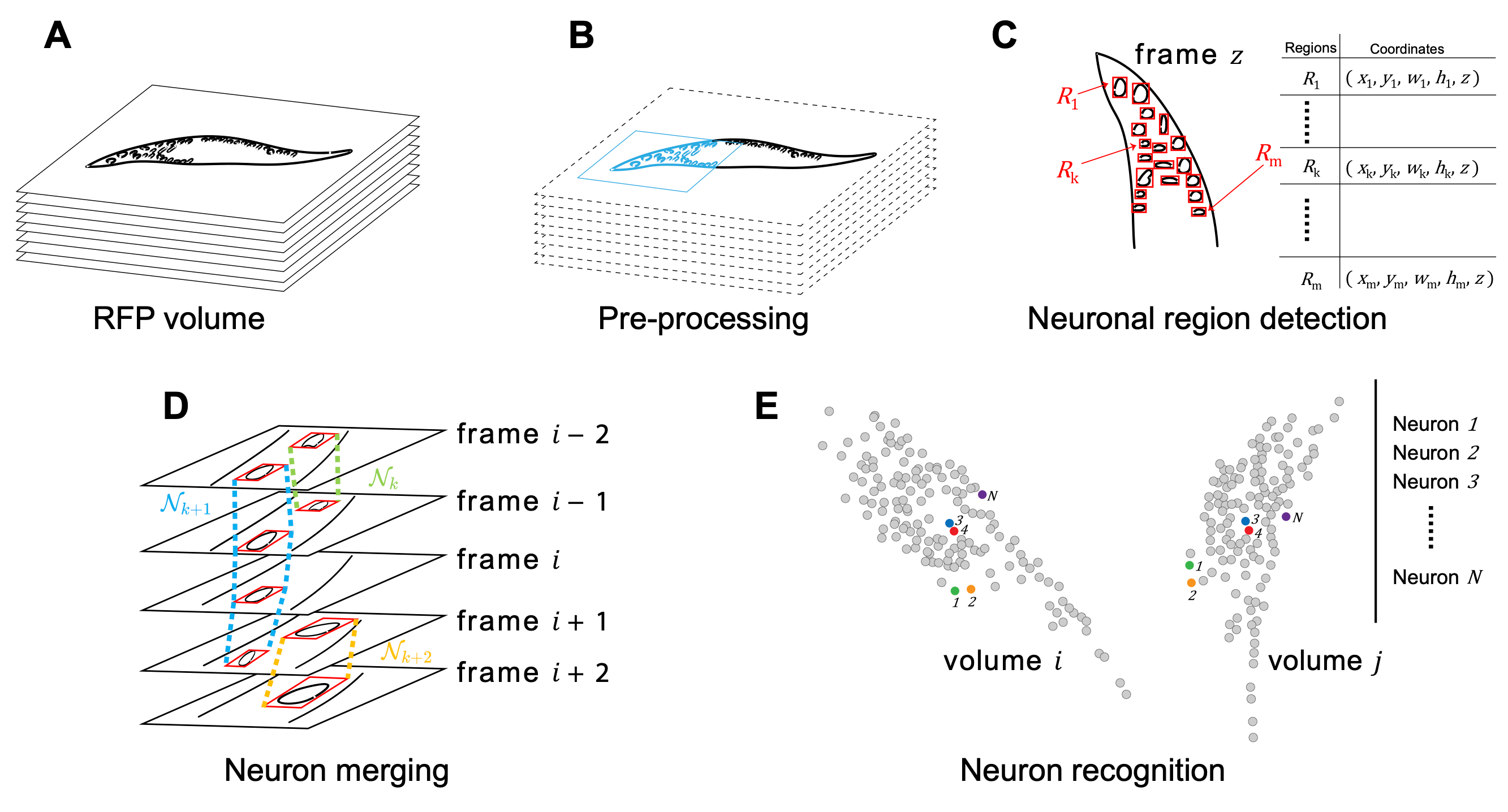
Advanced volumetric imaging methods and genetically encoded activity indicators have permitted a comprehensive characterization of whole brain activity at single neuron resolution in \textit{Caenorhabditis elegans}. The constant motion and deformation of the nematode nervous system, however, impose a great challenge for consistent identification of densely packed neurons in a behaving animal. Here, we propose a cascade solution for long-term and rapid recognition of head ganglion neurons in a freely moving \textit{C. elegans}. First, potential neuronal regions from a stack of fluorescence images are detected by a deep learning algorithm. Second, 2-dimensional neuronal regions are fused into 3-dimensional neuron entities. Third, by exploiting the neuronal density distribution surrounding a neuron and relative positional information between neurons, a multi-class artificial neural network transforms engineered neuronal feature vectors into digital neuronal identities. With a small number of training samples, our bottom-up approach is able to process each volume - $1024 \times 1024 \times 18$ in voxels - in less than 1 second and achieves an accuracy of $91\%$ in neuronal detection and above $80\%$ in neuronal tracking over a long video recording. Our work represents a step towards rapid and fully automated algorithms for decoding whole brain activity underlying naturalistic behaviors.
PDF Abstract