The Deep Generative Decoder: MAP estimation of representations improves modeling of single-cell RNA data
Learning low-dimensional representations of single-cell transcriptomics has become instrumental to its downstream analysis. The state of the art is currently represented by neural network models such as variational autoencoders (VAEs) which use a variational approximation of the likelihood for inference. We here present the Deep Generative Decoder (DGD), a simple generative model that computes model parameters and representations directly via maximum a posteriori (MAP) estimation. The DGD handles complex parameterized latent distributions naturally unlike VAEs which typically use a fixed Gaussian distribution, because of the complexity of adding other types. We first show its general functionality on a commonly used benchmark set, Fashion-MNIST. Secondly, we apply the model to multiple single-cell data sets. Here the DGD learns low-dimensional, meaningful and well-structured latent representations with sub-clustering beyond the provided labels. The advantages of this approach are its simplicity and its capability to provide representations of much smaller dimensionality than a comparable VAE.
PDF Abstract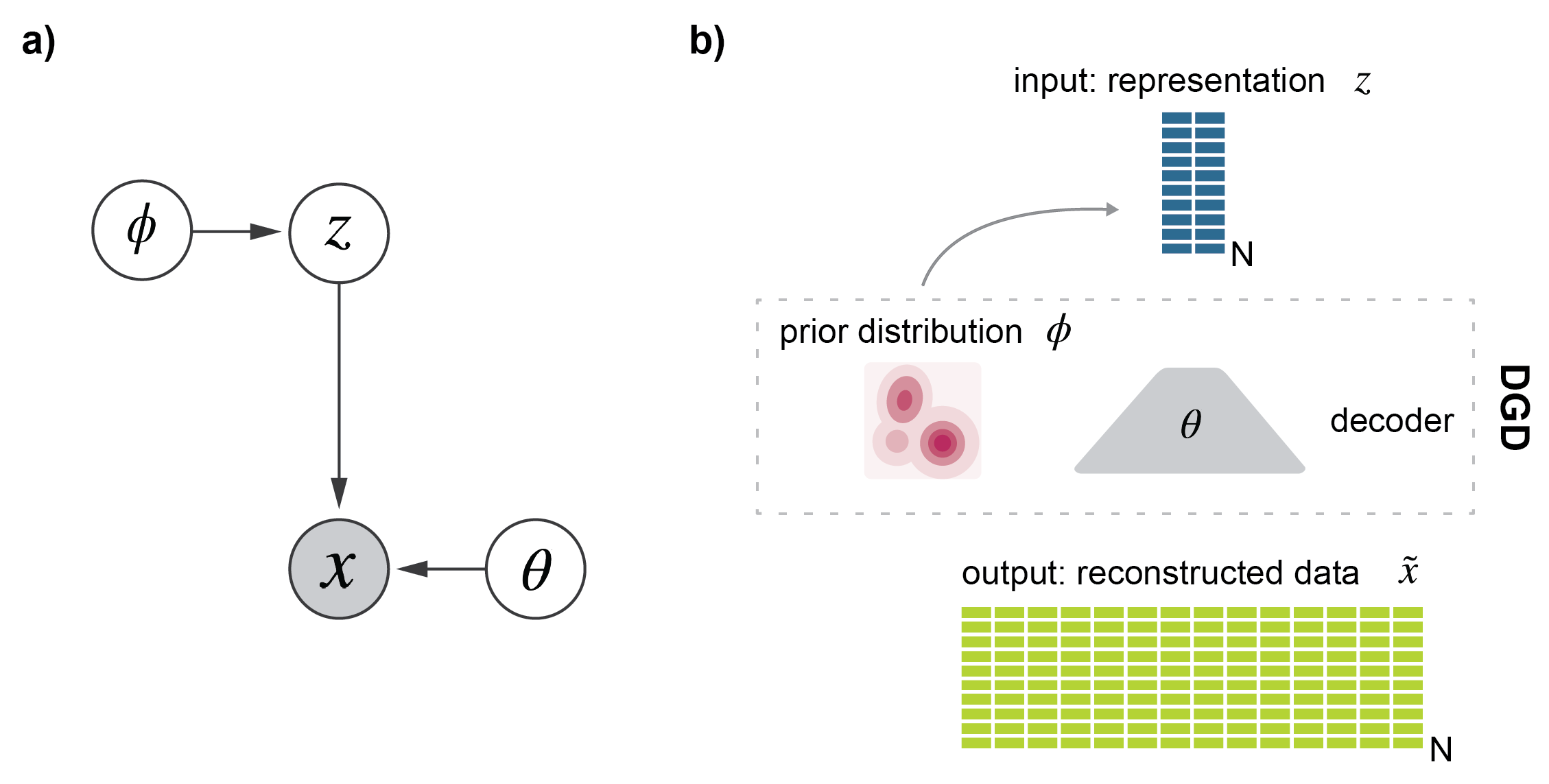




 Fashion-MNIST
Fashion-MNIST