Region-wise Loss for Biomedical Image Segmentation
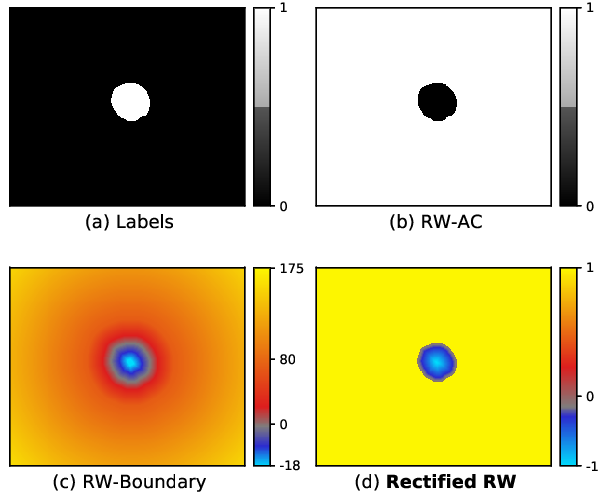
We propose Region-wise (RW) loss for biomedical image segmentation. Region-wise loss is versatile, can simultaneously account for class imbalance and pixel importance, and it can be easily implemented as the pixel-wise multiplication between the softmax output and a RW map. We show that, under the proposed RW loss framework, certain loss functions, such as Active Contour and Boundary loss, can be reformulated similarly with appropriate RW maps, thus revealing their underlying similarities and a new perspective to understand these loss functions. We investigate the observed optimization instability caused by certain RW maps, such as Boundary loss distance maps, and we introduce a mathematically-grounded principle to avoid such instability. This principle provides excellent adaptability to any dataset and practically ensures convergence without extra regularization terms or optimization tricks. Following this principle, we propose a simple version of boundary distance maps called rectified Region-wise (RRW) maps that, as we demonstrate in our experiments, achieve state-of-the-art performance with similar or better Dice coefficients and Hausdorff distances than Dice, Focal, weighted Cross entropy, and Boundary losses in three distinct segmentation tasks. We quantify the optimization instability provided by Boundary loss distance maps, and we empirically show that our RRW maps are stable to optimize. The code to run all our experiments is publicly available at: https://github.com/jmlipman/RegionWiseLoss.
PDF Abstract