A step toward a reinforcement learning de novo genome assembler
De novo genome assembly is a relevant but computationally complex task in genomics. Although de novo assemblers have been used successfully in several genomics projects, there is still no 'best assembler', and the choice and setup of assemblers still rely on bioinformatics experts. Thus, as with other computationally complex problems, machine learning may emerge as an alternative (or complementary) way for developing more accurate and automated assemblers. Reinforcement learning has proven promising for solving complex activities without supervision - such games - and there is a pressing need to understand the limits of this approach to 'real' problems, such as the DFA problem. This study aimed to shed light on the application of machine learning, using reinforcement learning (RL), in genome assembly. We expanded upon the sole previous approach found in the literature to solve this problem by carefully exploring the learning aspects of the proposed intelligent agent, which uses the Q-learning algorithm, and we provided insights for the next steps of automated genome assembly development. We improved the reward system and optimized the exploration of the state space based on pruning and in collaboration with evolutionary computing. We tested the new approaches on 23 new larger environments, which are all available on the internet. Our results suggest consistent performance progress; however, we also found limitations, especially concerning the high dimensionality of state and action spaces. Finally, we discuss paths for achieving efficient and automated genome assembly in real scenarios considering successful RL applications - including deep reinforcement learning.
PDF Abstract
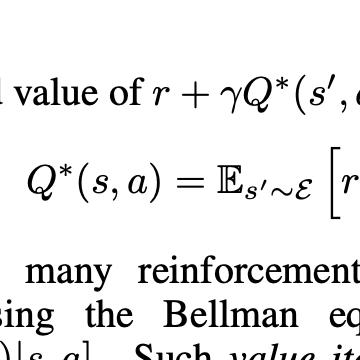

 OpenAI Gym
OpenAI Gym