Tracking by weakly-supervised learning and graph optimization for whole-embryo C. elegans lineages
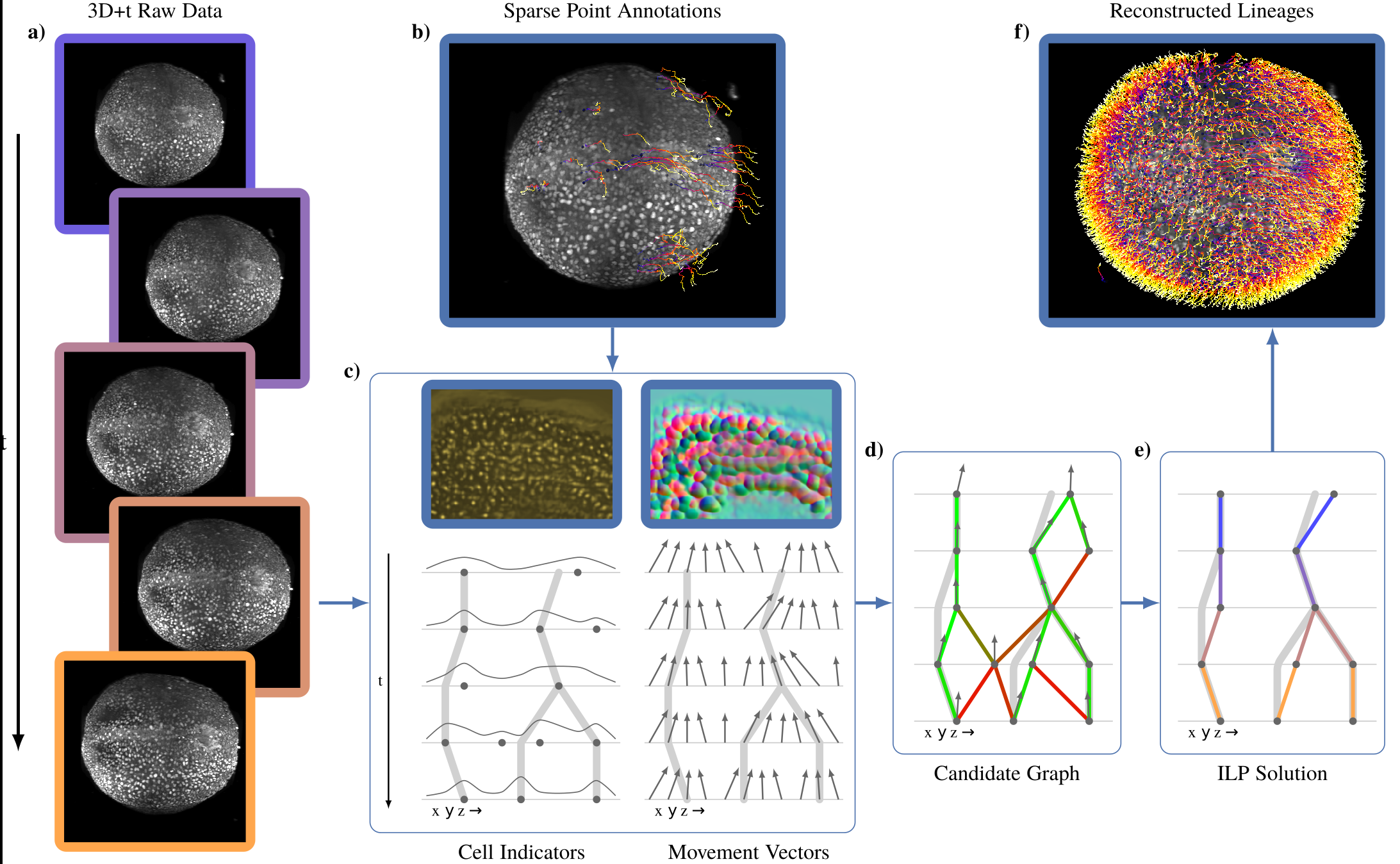
Tracking all nuclei of an embryo in noisy and dense fluorescence microscopy data is a challenging task. We build upon a recent method for nuclei tracking that combines weakly-supervised learning from a small set of nuclei center point annotations with an integer linear program (ILP) for optimal cell lineage extraction. Our work specifically addresses the following challenging properties of C. elegans embryo recordings: (1) Many cell divisions as compared to benchmark recordings of other organisms, and (2) the presence of polar bodies that are easily mistaken as cell nuclei. To cope with (1), we devise and incorporate a learnt cell division detector. To cope with (2), we employ a learnt polar body detector. We further propose automated ILP weights tuning via a structured SVM, alleviating the need for tedious manual set-up of a respective grid search. Our method outperforms the previous leader of the cell tracking challenge on the Fluo-N3DH-CE embryo dataset. We report a further extensive quantitative evaluation on two more C. elegans datasets. We will make these datasets public to serve as an extended benchmark for future method development. Our results suggest considerable improvements yielded by our method, especially in terms of the correctness of division event detection and the number and length of fully correct track segments. Code: https://github.com/funkelab/linajea
PDF Abstract