Unleashing the Potential of SAM for Medical Adaptation via Hierarchical Decoding
The Segment Anything Model (SAM) has garnered significant attention for its versatile segmentation abilities and intuitive prompt-based interface. However, its application in medical imaging presents challenges, requiring either substantial training costs and extensive medical datasets for full model fine-tuning or high-quality prompts for optimal performance. This paper introduces H-SAM: a prompt-free adaptation of SAM tailored for efficient fine-tuning of medical images via a two-stage hierarchical decoding procedure. In the initial stage, H-SAM employs SAM's original decoder to generate a prior probabilistic mask, guiding a more intricate decoding process in the second stage. Specifically, we propose two key designs: 1) A class-balanced, mask-guided self-attention mechanism addressing the unbalanced label distribution, enhancing image embedding; 2) A learnable mask cross-attention mechanism spatially modulating the interplay among different image regions based on the prior mask. Moreover, the inclusion of a hierarchical pixel decoder in H-SAM enhances its proficiency in capturing fine-grained and localized details. This approach enables SAM to effectively integrate learned medical priors, facilitating enhanced adaptation for medical image segmentation with limited samples. Our H-SAM demonstrates a 4.78% improvement in average Dice compared to existing prompt-free SAM variants for multi-organ segmentation using only 10% of 2D slices. Notably, without using any unlabeled data, H-SAM even outperforms state-of-the-art semi-supervised models relying on extensive unlabeled training data across various medical datasets. Our code is available at https://github.com/Cccccczh404/H-SAM.
PDF Abstract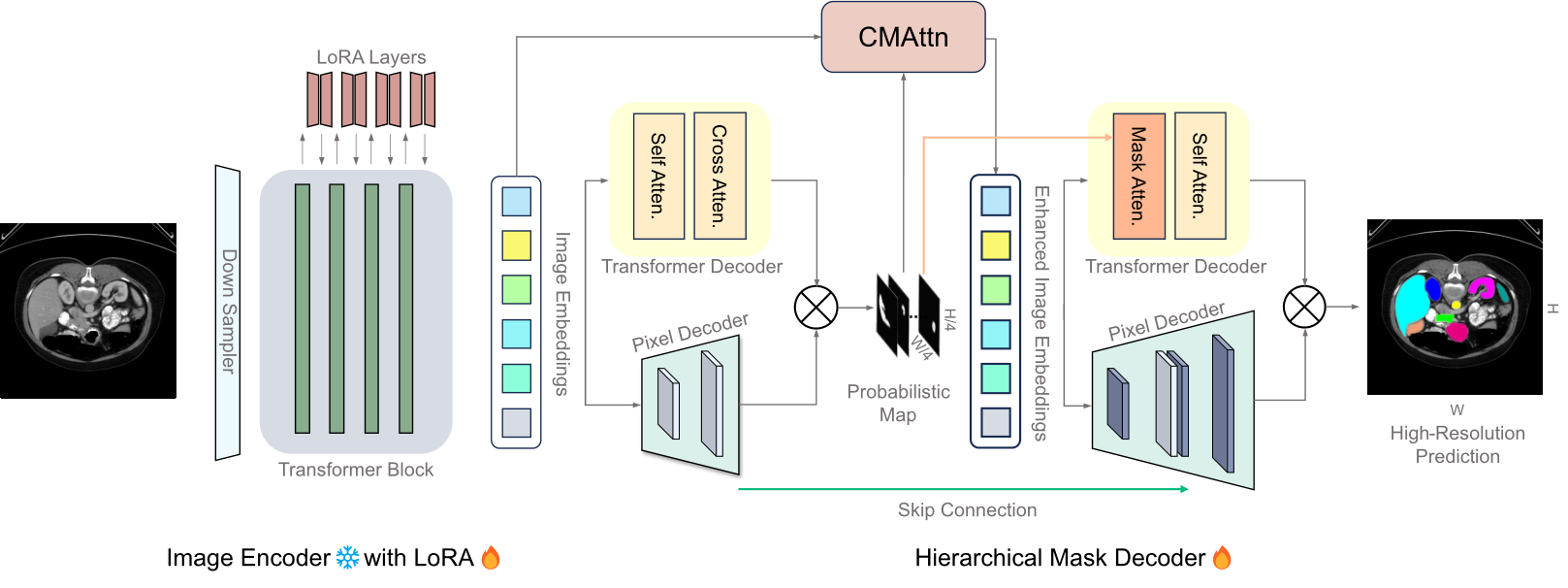




 PROMISE12
PROMISE12