Encoder-Decoder Architectures for Clinically Relevant Coronary Artery Segmentation
Coronary X-ray angiography is a crucial clinical procedure for the diagnosis and treatment of coronary artery disease, which accounts for roughly 16% of global deaths every year. However, the images acquired in these procedures have low resolution and poor contrast, making lesion detection and assessment challenging. Accurate coronary artery segmentation not only helps mitigate these problems, but also allows the extraction of relevant anatomical features for further analysis by quantitative methods. Although automated segmentation of coronary arteries has been proposed before, previous approaches have used non-optimal segmentation criteria, leading to less useful results. Most methods either segment only the major vessel, discarding important information from the remaining ones, or segment the whole coronary tree based mostly on contrast information, producing a noisy output that includes vessels that are not relevant for diagnosis. We adopt a better-suited clinical criterion and segment vessels according to their clinical relevance. Additionally, we simultaneously perform catheter segmentation, which may be useful for diagnosis due to the scale factor provided by the catheter's known diameter, and is a task that has not yet been performed with good results. To derive the optimal approach, we conducted an extensive comparative study of encoder-decoder architectures trained on a combination of focal loss and a variant of generalized dice loss. Based on the EfficientNet and the UNet++ architectures, we propose a line of efficient and high-performance segmentation models using a new decoder architecture, the EfficientUNet++, whose best-performing version achieved average dice scores of 0.8904 and 0.7526 for the artery and catheter classes, respectively, and an average generalized dice score of 0.9234.
PDF Abstract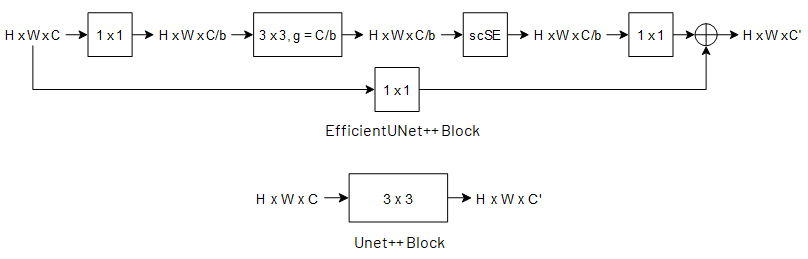


 100DOH
100DOH