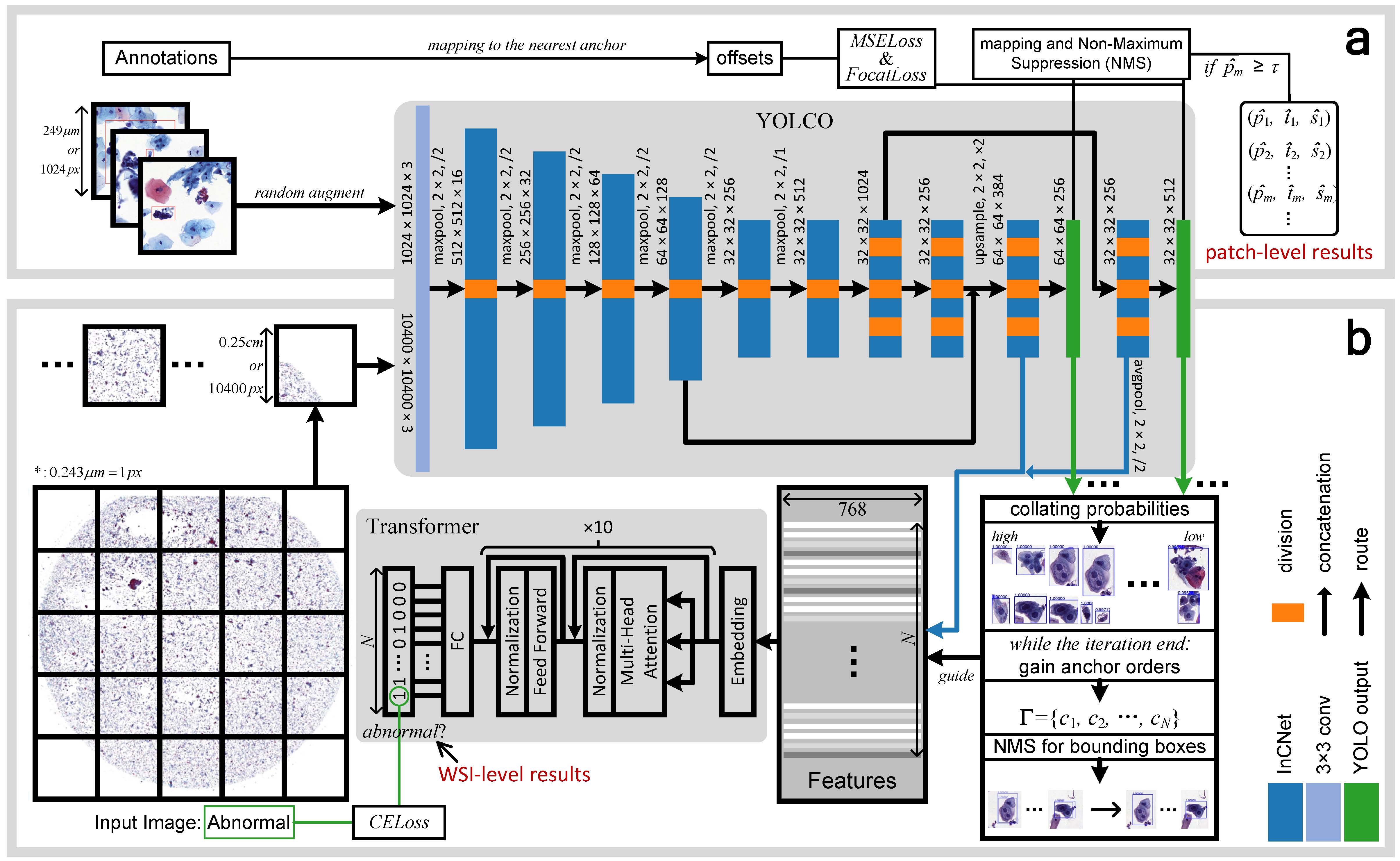
An Efficient Cervical Whole Slide Image Analysis Framework Based on Multi-scale Semantic and Location Deep Features
Digital gigapixel whole slide image (WSI) is widely used in clinical diagnosis, and automated WSI analysis is key for computer-aided diagnosis. Currently, analyzing the integrated descriptor of probabilities or feature maps from massive local patches encoded by ResNet classifier is the main manner for WSI-level prediction. Feature representations of the sparse and tiny lesion cells in cervical slides, however, are still challenging, while the unused location representations are available to supply the semantics classification. This study designs a novel and efficient framework with a new module InCNet constructed lightweight model YOLCO (You Only Look Cytology Once). It directly extracts feature inside the single cell (cluster) instead of the traditional way that from image tile with a fixed size. The InCNet (Inline Connection Network) enriches the multi-scale connectivity without efficiency loss. The proposal allows the input size enlarged to megapixel that can stitch the WSI by the average repeats decreased from $10^3\sim10^4$ to $10^1\sim10^2$ for collecting features and predictions at two scales. Based on Transformer for classifying the integrated multi-scale multi-task WSI features, the experimental results appear $0.872$ AUC score better than the best conventional model on our dataset ($n$=2,019) from four scanners. The code is available at https://github.com/Chrisa142857/You-Only-Look-Cytopathology-Once , where the deployment version has the speed $\sim$70 s/WSI.
PDF Abstract