TransNorm: Transformer Provides a Strong Spatial Normalization Mechanism for a Deep Segmentation Model
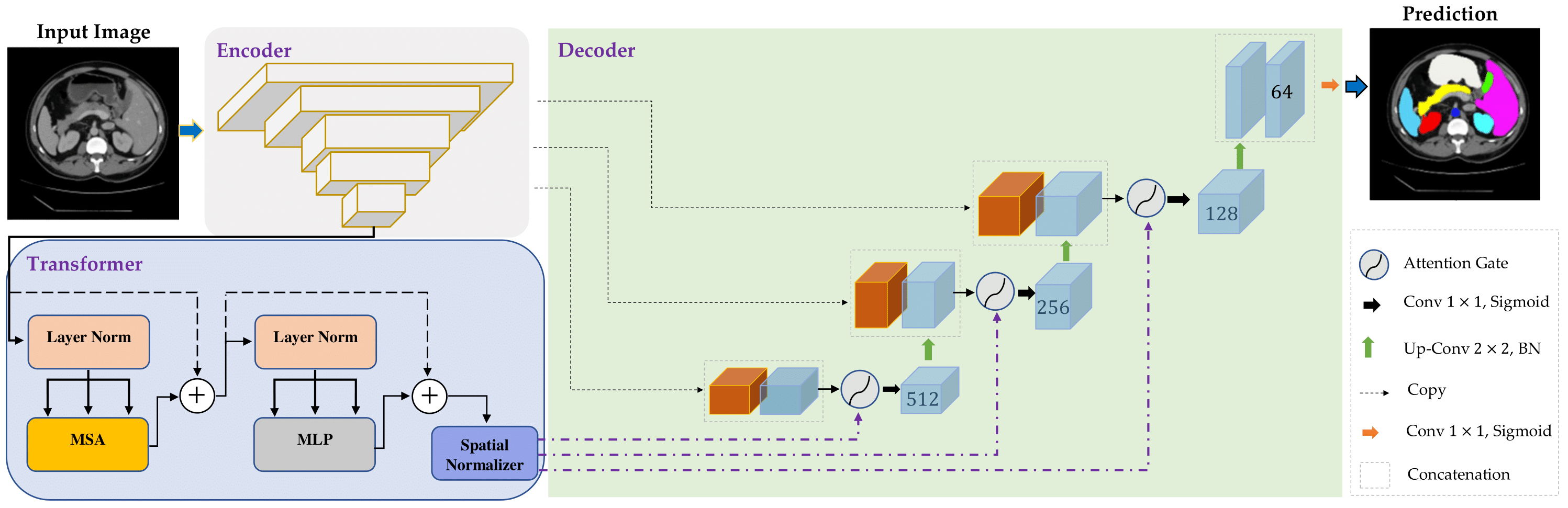
In the past few years, convolutional neural networks (CNNs), particularly U-Net, have been the prevailing technique in the medical image processing era. Specifically, the seminal U-Net, as well as its alternatives, have successfully managed to address a wide variety of medical image segmentation tasks. However, these architectures are intrinsically imperfect as they fail to exhibit long-range interactions and spatial dependencies leading to a severe performance drop in the segmentation of medical images with variable shapes and structures. Transformers, preliminary proposed for sequence-to-sequence prediction, have arisen as surrogate architectures to precisely model global information assisted by the self-attention mechanism. Despite being feasibly designed, utilizing a pure Transformer for image segmentation purposes can result in limited localization capacity stemming from inadequate low-level features. Thus, a line of research strives to design robust variants of Transformer-based U-Net. In this paper, we propose Trans-Norm, a novel deep segmentation framework which concomitantly consolidates a Transformer module into both encoder and skip-connections of the standard U-Net. We argue that the expedient design of skip-connections can be crucial for accurate segmentation as it can assist in feature fusion between the expanding and contracting paths. In this respect, we derive a Spatial Normalization mechanism from the Transformer module to adaptively recalibrate the skip connection path. Extensive experiments across three typical tasks for medical image segmentation demonstrate the effectiveness of TransNorm. The codes and trained models are publicly available at https://github.com/rezazad68/transnorm.
PDF Abstract